CRISPR-Cas9 system discovery has revolutionized the genetic field by promoting fast and efficient precise genome editing. Only in less than a decade after CRISPR-Cas9 characterization, up to a dozen of clinical trials have been proposed. CRISPR-Cas9 system is mainly used to induce DNA targeted modification at the nucleotidic level. But recently, two teams of researchers have used CRISPR to trigger entire chromosome deletion and paved the way for what they called “the chromosomal therapies”. For that, they induced several double-strand breaks (DSBs) all along the targeted chromosome and that lead to the full deletion of this last. Using this approach, Adikusuma et al.1 achieved to create mouse model with Turner syndrome by targeting the chromosome Y in mice embryos. Zuo et al.2 also achieved the correction of Down syndrome by targeting the chromosome 21 in human iPSCs.
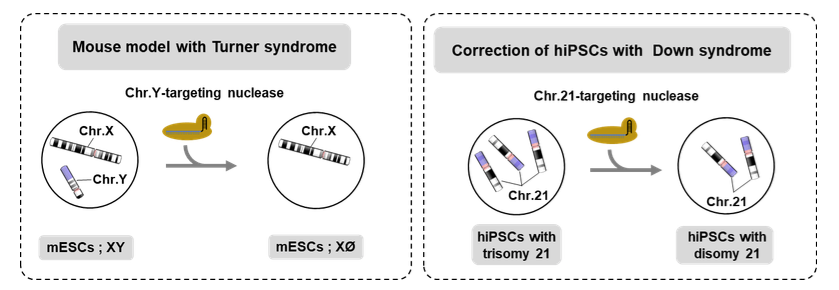
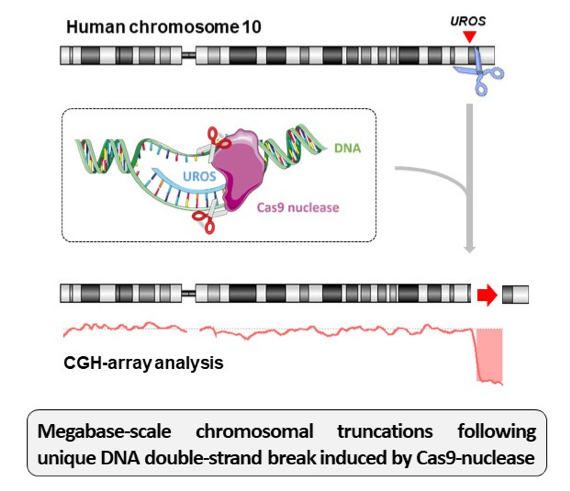
But in a way, these works have warned us on how toxic can be several DSBs on chromosomal integrity. So, in sight of gene therapy, we asked ourselves if a single DSB can impair the targeted chromosome3. As classical analysis of nuclease-induced outcomes relies on PCR that cannot detect chromosomal abnormalities, we decided to carry out a cytogenetic analysis of the Cas9-induced events. As gene editing model, we have targeted UROS gene (implied in congenital erythropoietic porphyria) that is located in the long arm of chromosome 10. Using a couple of subtelomeric FISH probes, we have observed, in some nuclei, a signal loss of the probe normally located next to the long-arm telomere. It suggests Cas9-nuclease can impair chromosomal integrity by terminal truncation of the chromosome. Array-CGH confirmed the terminal truncation phenotype and showed it started exactly at the targeted locus. This has been observed in immortalized fibroblasts knocked-out for TP53. However, one crucial question regarding biosafety clinical trials is to know if terminal truncations can occur in primary cells. Fortunately, we did not observed a chromosomal abnormalities rate enough sufficient to be detected by FISH in primary fibroblasts. However, as it can’t be excluded, we are developing an alternative approach excluding the use of a DSB. For that, we used the Cas9D10A-nickase that induce a single-strand break (SSB), instead of DSB, and co-supplied the cells with a single-strand desoxynucleotide (ssODN) to able homology-directed repair (HDR). This approach allows to avoid both nuclease-triggered indels and terminal truncation, offering a safer way to perform gene editing.
Finally, I would like to thanks SFTCG for setting up this thematic day on viral vector that took place at the Hospital La Pitié-Salepêtrière (Paris). Letting young researchers presenting their thesis work is great opportunity and I’m honoured to have been chosen for the best oral communications. I also acknowledge the ESGCT for sponsoring this prize and supporting and enhancing cooperation of gene and cell therapy field all around the scientific community.
1- Targeted Deletion of an Entire Chromosome Using CRISPR/Cas9. Adikusuma F et al., Mol Ther., August 2017
2- CRISPR/Cas9-mediated targeted chromosome elimination. Zuo E. et al. – Genome Biol., November 2017
3- CRISPR-Cas9 genome editing induces megabase-scale chromosomal truncations. Cullot G. et al. – Nat Commun., March 2019